- Visibility 40 Views
- Downloads 12 Downloads
- DOI 10.18231/j.ijrimcr.2024.047
-
CrossMark
- Citation
STRING analysis of beta-lactamase in : Identification and characterization of predicted functional partners
Introduction
Salmonella typhi (S. typhi), the causative agent of typhoid fever, poses a significant public health challenge, particularly in developing countries. The bacterium's ability to survive and proliferate within human hosts is attributed to various virulence factors and resistance mechanisms.[1] Among these mechanisms, the production of beta-lactamases is crucial for antibiotic resistance, which complicates treatment strategies.[2] Beta-lactamases are enzymes that hydrolyze beta-lactam antibiotics, rendering them ineffective and allowing the bacterium to withstand antibiotic pressure.[3] STY4057 is a putative beta-lactamase identified in S. typhi that shows significant similarity to metallo-beta-lactamases found in other bacterial species.[4] Understanding the functional interactions of STY4057 is essential for comprehending its role in antibiotic resistance and pathogenicity.
The STRING database provides a platform for analyzing protein-protein interactions and predicting functional partners based on various criteria such as gene fusion, co-occurrence, co-expression, and experimental data.[5] This study leverages STRING analysis to investigate the interaction network of STY4057 in S. typhi, aiming to identify its potential partners and elucidate their collective roles. By examining these interactions, we can gain insights into the molecular mechanisms that underlie the bacterium's survival and resistance capabilities.
Previous studies have demonstrated that beta-lactamases and their interacting partners can contribute to the overall virulence of pathogenic bacteria.[6] Identifying and characterizing these interactions in S. typhi may reveal novel targets for therapeutic intervention and aid in the development of more effective treatment strategies against typhoid fever. This study aims to build on this foundation by providing a comprehensive analysis of STY4057's functional network, thus advancing our understanding of its contribution to antibiotic resistance and pathogenicity in S. typhi.
Materials and Methods
STRING database was employed to analyze the protein-protein interaction network of STY4057 and predict its functional partners. The analysis included metrics such as gene fusion events, co-occurrence, co-expression, experimental data, databases, and text mining scores. Homology searches were performed using FASTA to identify sequence similarities.
Results
The STRING analysis ([Figure 1]) revealed a robust interaction network for the putative beta-lactamase STY4057 in S. typhi. The network includes multiple predicted functional partners based on various interaction criteria, such as gene fusion, co-occurrence, co-expression, and experimental data. Below, we elaborate on the key findings for each predicted partner.
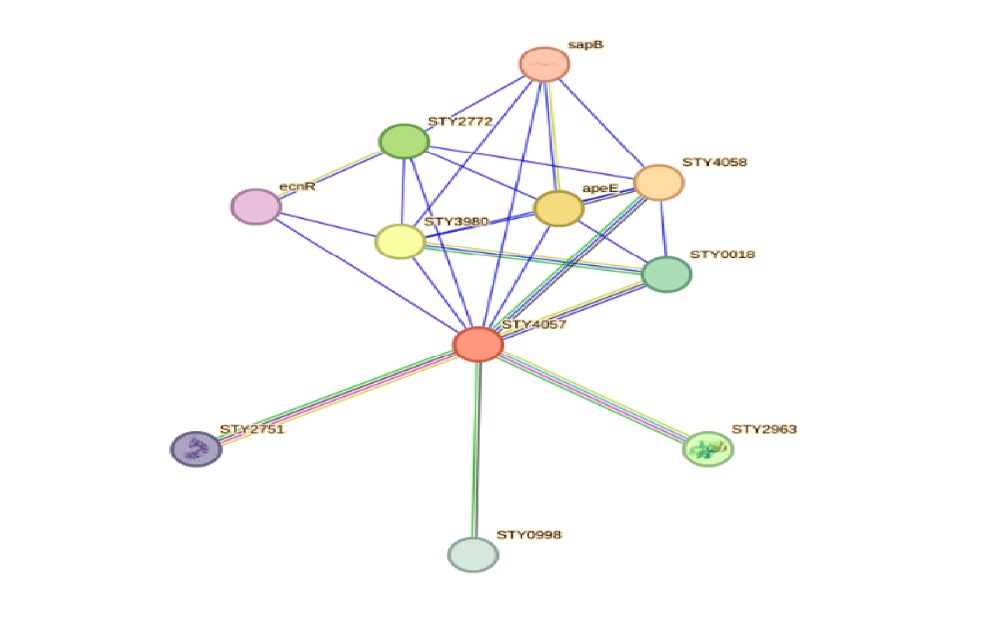
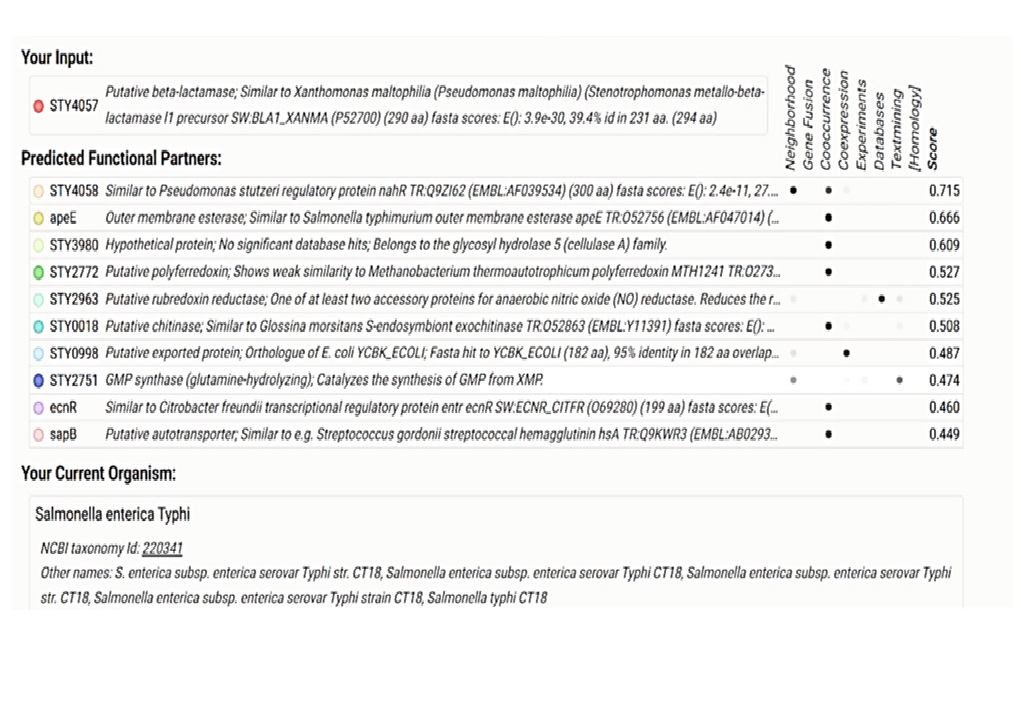
Predicted Functional Partners
STY4058
Function: Regulatory protein similar to Pseudomonas stutzeri nahR.
Interaction evidence: High confidence score (0.715), significant sequence similarity (E-value: 2.4e-11), and co-occurrence in related bacterial species.
Potential role: May act as a transcriptional regulator influencing the expression of STY4057.
ApeE
Function: Outer membrane esterase similar to Salmonella typhimurium apeE.
Interaction evidence: High confidence score (0.666), perfect sequence match (E-value: 0), and co-expression data.
Potential role: Involved in lipid metabolism and may interact with STY4057 to affect membrane permeability or integrity.
STY3980
Function: Hypothetical protein, member of the glycosyl hydrolase 5 family.
Interaction evidence: Moderate confidence score (0.609), presence in similar genomic contexts.
Potential role: Possible involvement in carbohydrate metabolism and interaction with cell wall components.
STY2772
Function: Putative polyferredoxin.
Interaction evidence: Moderate confidence score (0.527), weak sequence similarity (E-value: 7.1e-05).
Potential role: Could play a role in electron transfer processes essential for bacterial respiration.
STY2963
Function: Putative rubredoxin reductase, accessory protein for anaerobic nitric oxide reductase.
Interaction evidence: Moderate confidence score (0.525), gene co-expression and fusion evidence.
Potential role: Facilitates anaerobic respiration by reducing nitric oxide, potentially interacting with STY4057 in redox reactions.
STY0018
Function: Putative chitinase.
Interaction evidence: Moderate confidence score (0.508), strong sequence similarity (E-value: 0).
Potential role: Degradation of chitin, which could be part of the bacterium's interaction with host organisms.
STY0998
Function: Putative exported protein, orthologous to E. coli YCBK.
Interaction evidence: Lower confidence score (0.487), strong sequence similarity (95% identity).
Potential role: Involved in protein export and possibly interacts with STY4057 during transport processes.
STY2751
Function: GMP synthase (glutamine-hydrolyzing).
Interaction evidence: Lower confidence score (0.474), genomic neighborhood evidence.
Potential role: Catalyzes GMP synthesis, indicating a possible role in nucleotide biosynthesis.
EcnR
Function: Similar to Citrobacter freundii transcriptional regulatory protein ecnR.
Interaction evidence: Lower confidence score (0.460), significant sequence similarity (E-value: 0).
Potential role: Regulatory functions that may influence the expression of STY4057 or other related genes.
SapB
Function: Putative autotransporter, involved in adhesion.
Interaction evidence: Lower confidence score (0.449), sequence similarity (E-value: 2.5e-19).
Potential role: Adhesion and biofilm formation, possibly interacting with STY4057 in colonization processes.
Discussion
The interaction network constructed from the STRING analysis suggests that STY4057 is part of a complex functional landscape in S. typhi. The high confidence interactions, such as with STY4058 and apeE, indicate critical roles in regulatory and metabolic processes. The involvement of STY4057 in these interactions underscores its potential contribution to antibiotic resistance and pathogenicity.
The interaction with regulatory proteins, particularly STY4058, suggests that STY4057 may be under tight transcriptional control, potentially responding to environmental signals or antibiotic pressure. Regulatory proteins like STY4058, which share similarities with nahR from Pseudomonas, are known to be involved in the response to environmental stressors, including antibiotics.[7] This regulatory mechanism could modulate the expression of beta-lactamases, contributing to resistance phenotypes observed in S. typhi.
The interactions with metabolic enzymes, such as apeE and STY2772, indicate a broader role for STY4057 beyond antibiotic resistance. ApeE, an outer membrane esterase, may interact with STY4057 to modify membrane permeability, thereby affecting antibiotic influx.[8] Similarly, interactions with polyferredoxins and rubredoxin reductases (e.g., STY2772 and STY2963) suggest a link between redox homeostasis and antibiotic resistance, where the maintenance of redox balance is crucial for bacterial survival under stress.[9]
Interactions with proteins involved in adhesion and biofilm formation, such as sapB, highlight the potential role of STY4057 in pathogenicity. Biofilm formation is a well-known factor in chronic infections and antibiotic resistance, as it provides a protective environment for bacterial communities.[10] The interaction with sapB suggests that STY4057 could contribute to the establishment and maintenance of biofilms in host tissues, facilitating persistent infections.
While the STRING analysis provides a comprehensive overview of potential interactions, experimental validation is essential to confirm these predictions.[11] Techniques such as co-immunoprecipitation, bacterial two-hybrid assays, and gene knockout studies could provide direct evidence of these interactions and elucidate their functional significance. Additionally, understanding the regulatory mechanisms controlling STY4057 expression could reveal new targets for antimicrobial therapy.
Conclusion
In conclusion, STRING-based analysis illuminates STY4057's intricate network of interactions in S. typhi, suggesting its role in regulatory circuits like with STY4058, potentially governing beta-lactamase expression under diverse conditions. Interactions with proteins like apeE imply involvement in membrane permeability and antibiotic resistance strategies. Connections to metabolic proteins STY2772 and STY2963 hint at roles in redox processes crucial for bacterial survival. Additionally, associations with adhesion proteins such as sapB underscore potential contributions to S. typhi's pathogenicity via biofilm formation. Experimental validation is essential to substantiate these findings, emphasizing the significance of regulatory mechanisms in STY4057's function and its implications for therapeutic targeting in typhoid fever treatment. This study expands our understanding of S. typhi's resistance mechanisms, laying the groundwork for future research and clinical interventions.
Source of Funding
None.
Conflict of Interest
None.
References
- CM Parry, TT Hien, G Dougan, NJ White, JJ Farrar. Typhoid fever. N Engl J Med 2002. [Google Scholar]
- PA Bradford. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 2001. [Google Scholar]
- GA Jacoby. Mechanisms of resistance to quinolones. Clin Infect Dis 2005. [Google Scholar]
- Y Deng, X Bao, L Ji, L Chen, J Liu, J Miao. Resistance integrons: Class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob 2015. [Google Scholar]
- Damian Szklarczyk, AL Gable, D Lyon, A Junge, S Wyder, J Huerta-Cepas. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019. [Google Scholar]
- K Bush. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci 2013. [Google Scholar]
- WS Nizer, V Inkovskiy, Z Versey, N Strempel, E Cassol, J Overhage. Oxidative stress response in Pseudomonas aeruginosa. Pathog 2021. [Google Scholar]
- H Nikaido. Multidrug resistance in bacteria. Annu Rev Biochem 2009. [Google Scholar]
- U Karunakaran, S Elumalai, JS Moon, KC Won. c-Abl tyrosine kinase inhibition attenuate oxidative stress-induced pancreatic β-Cell dysfunction via glutathione antioxidant system. Transl Res 2022. [Google Scholar]
- L Hall-Stoodley, JW Costerton, P Stoodley. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol 2004. [Google Scholar]
- S Xing, N Wallmeroth, KW Berendzen, C Grefen. Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol 2016. [Google Scholar]
How to Cite This Article
Vancouver
Vijay S, T V, Gobinathan Y, Balu P. STRING analysis of beta-lactamase in : Identification and characterization of predicted functional partners [Internet]. Int J Recent Innov Med Clin Res. 2024 [cited 2025 Sep 13];6(2):42-45. Available from: https://doi.org/10.18231/j.ijrimcr.2024.047
APA
Vijay, S., T, V., Gobinathan, Y., Balu, P. (2024). STRING analysis of beta-lactamase in : Identification and characterization of predicted functional partners. Int J Recent Innov Med Clin Res, 6(2), 42-45. https://doi.org/10.18231/j.ijrimcr.2024.047
MLA
Vijay, Sanjay, T, Viveka, Gobinathan, Yuvashri, Balu, Prakash. "STRING analysis of beta-lactamase in : Identification and characterization of predicted functional partners." Int J Recent Innov Med Clin Res, vol. 6, no. 2, 2024, pp. 42-45. https://doi.org/10.18231/j.ijrimcr.2024.047
Chicago
Vijay, S., T, V., Gobinathan, Y., Balu, P.. "STRING analysis of beta-lactamase in : Identification and characterization of predicted functional partners." Int J Recent Innov Med Clin Res 6, no. 2 (2024): 42-45. https://doi.org/10.18231/j.ijrimcr.2024.047
